Overview
The Fazal lab aims to identify the RNA repertoire of various subcellular locations and uncover mechanisms employed by cells to localize these RNAs. We use APEX-seq-based proximity biotinylation to profile these RNAs. APEX, an ascorbate peroxidase, when targeted to various subcellular compartments biotinylates proximal RNAs, which can be later enriched and sequenced, thus providing the subcellular address of RNAs at a transcriptome-wide scale. We utilize a combination of experimental and computational approaches to understand the subcellular regulation of RNAs.
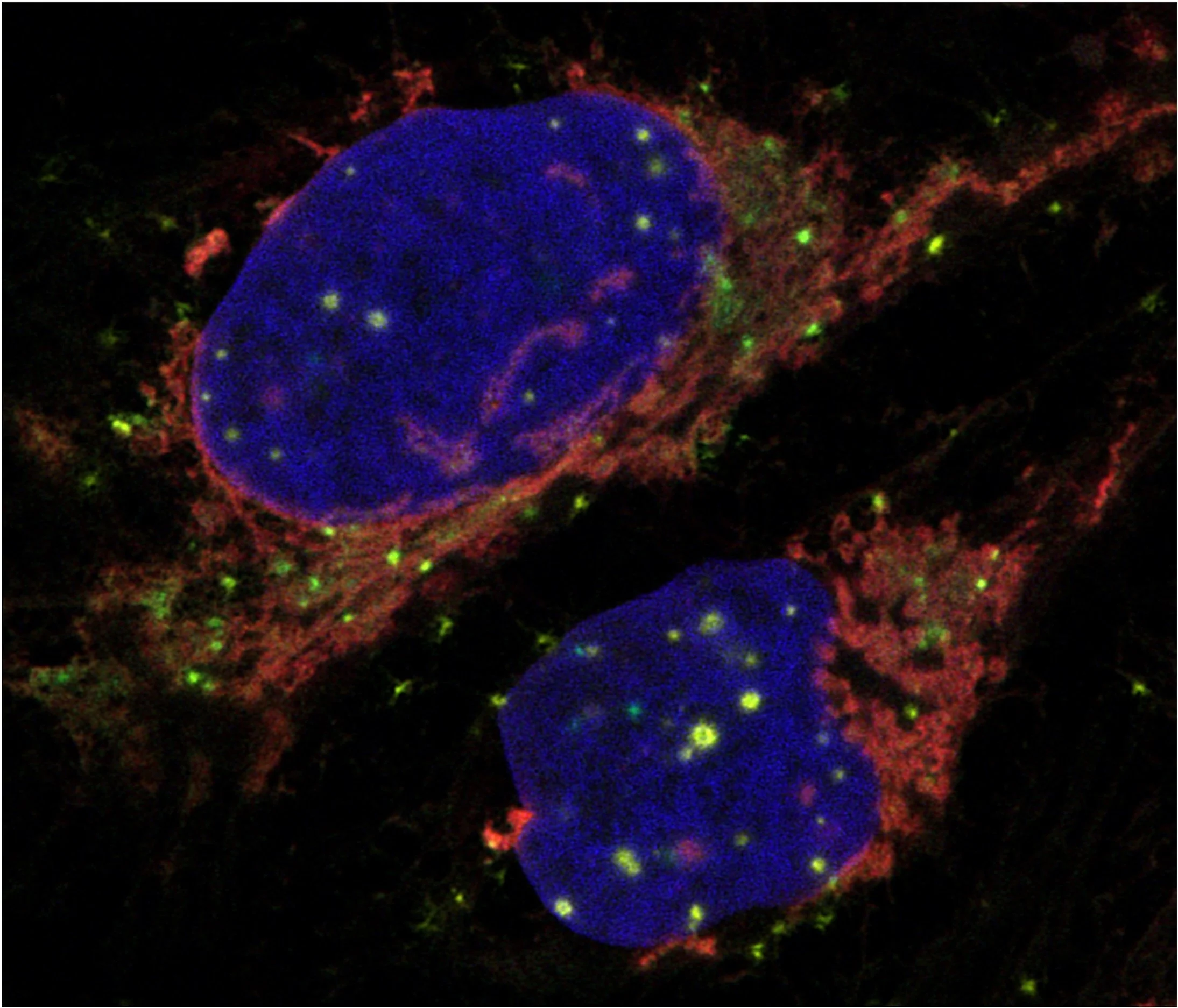
I.Mapping the Subcellular RNA Landscape
We previously identified the RNA landscape of nine subcellular compartments at transcriptome-wide scale. We continue to expand our approach to study novel and challenging subcellular locations such as nuclear membrane-less organelles, and expand to new cellular models.
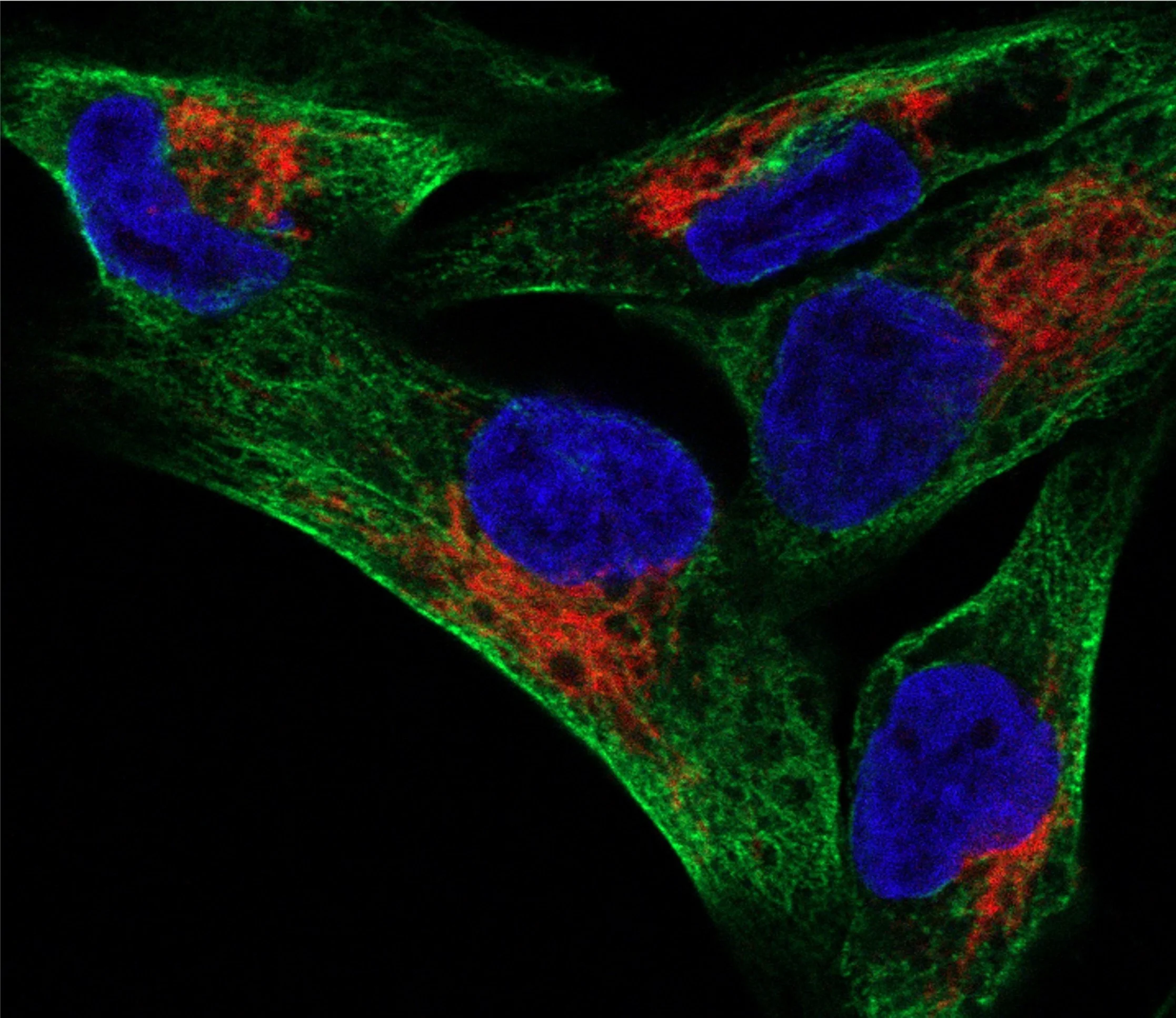
II. Investigating RNA-based Regulation of the Mitochondria
We discovered hundreds of transcripts coding for mitochondrial proteins localize to, and are translated at the surface of the mitochondria. Using APEX-seq, we study the regulation and translation of these RNAs.
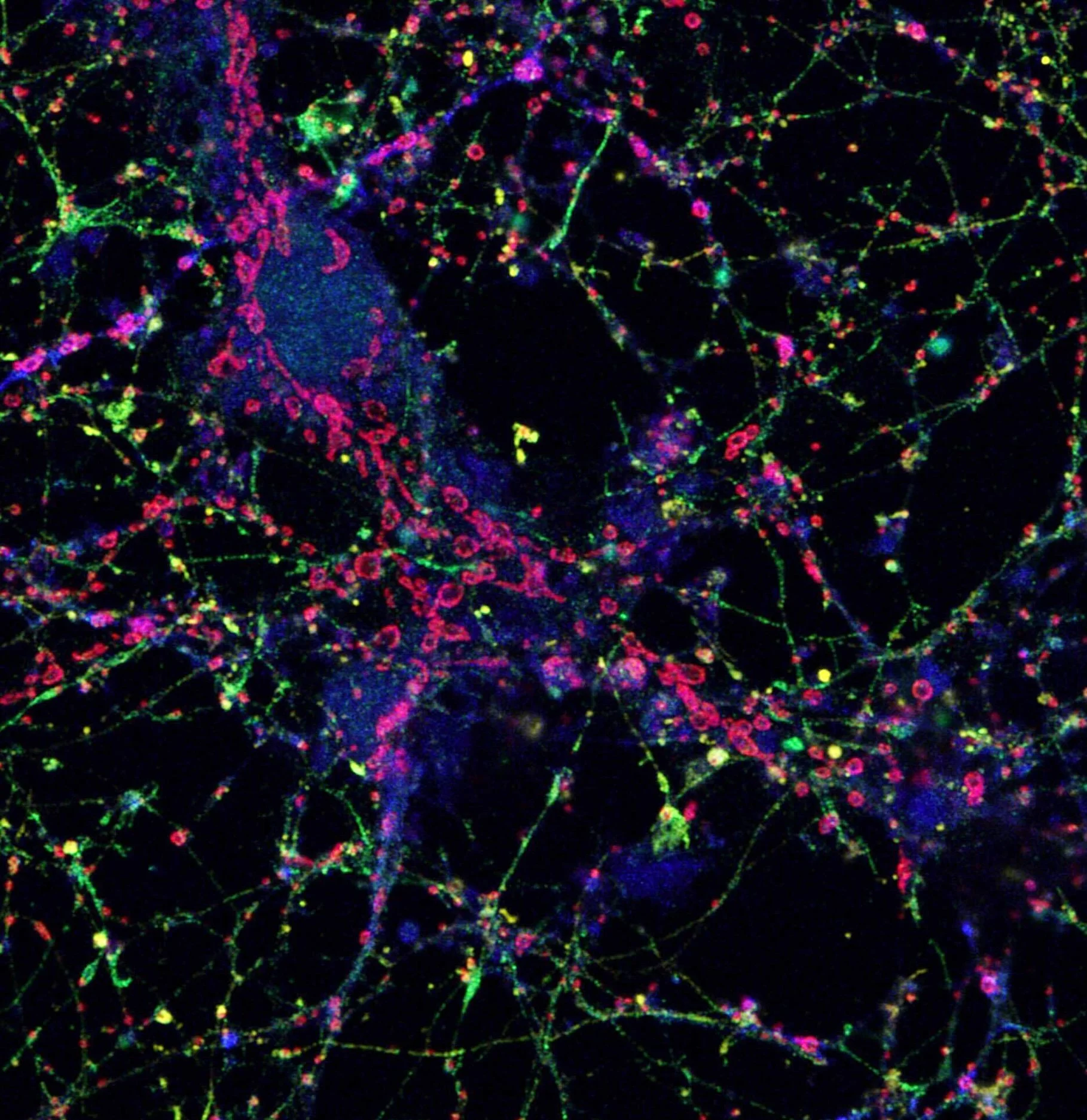
III. Identifying the Mechanisms of RNA Localization
We are currently working to unravel the mechanisms of subcellular RNA localization. To do so, we employ high-throughput assays to identify sequence motifs and their potential protein partners responsible for localizing RNAs. We also study the role of molecular motors in mediating the localization. Additionally, we develop computational models to derive features of RNAs localizing to a particular location.
IV. Investigating RNA Localization Defects in Disease models
We aim to translate our findings to disease models and identify the role of RNA mis-localization in disease progression. To this end, we introduce APEX constructs targeted to various subcellular locales in disease models such as triple-negative breast cancer and glioblastoma cells and profile the changes in RNA localization compared to normal state.
Featured Publications
RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus
Wu KE, Fazal FM, Parker KR, Zou J, Chang HY.
Cell Systems, 2020
Subcellular spatial transcriptomes: emerging frontier for understanding gene regulation
Fazal FM, Chang HY.
Cold Spring Harbor Symposium on Quantitative Biology, 2020
Atlas of Subcellular RNA Localization Revealed by APEX-seq
Fazal FM*, Han S*, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY.
Cell, 2019
RNA structure maps across mammalian cellular compartments
Sun L*, Fazal FM*, Li P*, Broughton JP, Lee B, Tang L, Huang W, Kool ET, Chang HY, Zhang QC.
Nature Structural and Molecular Biology (NSMB), 2019
Real-time observation of the initiation of RNA polymerase II transcription
Fazal FM*, Meng CA*, Murakami K*, Kornberg RD, Block SM.
Nature, 2015